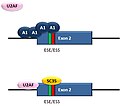
Alternative splicing
Alternative splicing is a regulated process during gene expression that results in a single gene coding for multiple proteins. This process is a key factor in the complexity of proteins in eukaryotes and is one of the main reasons why eukaryotes can have a relatively small number of genes but a much larger number of proteins.
Overview[edit]
In the process of transcription, the gene is copied into messenger RNA (mRNA). This mRNA then undergoes a process called splicing where introns (non-coding regions) are removed and exons (coding regions) are joined together. In alternative splicing, different combinations of exons are joined together to form different mRNAs, which are then translated into different proteins.
Mechanism[edit]
Alternative splicing can occur in several ways. The most common types are exon skipping, intron retention, mutually exclusive exons, and alternative 5' or 3' splice sites. The choice of which exons to include in the mRNA is determined by a complex set of regulatory signals in the DNA and RNA, as well as proteins and small RNAs in the cell.
Regulation[edit]
The regulation of alternative splicing is a complex process that involves numerous trans-acting factors and cis-acting elements. Trans-acting factors are proteins and small RNAs that bind to the mRNA and influence the splicing process. Cis-acting elements are sequences in the DNA or RNA that influence splicing.
Role in disease[edit]
Abnormal alternative splicing has been implicated in numerous diseases, including cancer, neurodegenerative diseases, and cardiovascular diseases. Understanding the mechanisms of alternative splicing and how it is regulated can therefore have important implications for the development of new treatments for these diseases.
See also[edit]
-
Alternative splicing
-
Alternative splicing
-
Alternative splicing
-
Alternative splicing
-
Alternative splicing
-
Alternative splicing
-
Alternative splicing
-
Alternative splicing
-
Alternative splicing
-
Alternative splicing
-
Alternative splicing
Ad. Transform your life with W8MD's Budget GLP-1 injections from $75


W8MD offers a medical weight loss program to lose weight in Philadelphia. Our physician-supervised medical weight loss provides:
- Weight loss injections in NYC (generic and brand names):
- Zepbound / Mounjaro, Wegovy / Ozempic, Saxenda
- Most insurances accepted or discounted self-pay rates. We will obtain insurance prior authorizations if needed.
- Generic GLP1 weight loss injections from $75 for the starting dose.
- Also offer prescription weight loss medications including Phentermine, Qsymia, Diethylpropion, Contrave etc.
NYC weight loss doctor appointmentsNYC weight loss doctor appointments
Start your NYC weight loss journey today at our NYC medical weight loss and Philadelphia medical weight loss clinics.
- Call 718-946-5500 to lose weight in NYC or for medical weight loss in Philadelphia 215-676-2334.
- Tags:NYC medical weight loss, Philadelphia lose weight Zepbound NYC, Budget GLP1 weight loss injections, Wegovy Philadelphia, Wegovy NYC, Philadelphia medical weight loss, Brookly weight loss and Wegovy NYC
|
WikiMD's Wellness Encyclopedia |
| Let Food Be Thy Medicine Medicine Thy Food - Hippocrates |
Medical Disclaimer: WikiMD is not a substitute for professional medical advice. The information on WikiMD is provided as an information resource only, may be incorrect, outdated or misleading, and is not to be used or relied on for any diagnostic or treatment purposes. Please consult your health care provider before making any healthcare decisions or for guidance about a specific medical condition. WikiMD expressly disclaims responsibility, and shall have no liability, for any damages, loss, injury, or liability whatsoever suffered as a result of your reliance on the information contained in this site. By visiting this site you agree to the foregoing terms and conditions, which may from time to time be changed or supplemented by WikiMD. If you do not agree to the foregoing terms and conditions, you should not enter or use this site. See full disclaimer.
Credits:Most images are courtesy of Wikimedia commons, and templates, categories Wikipedia, licensed under CC BY SA or similar.
Translate this page: - East Asian
中文,
日本,
한국어,
South Asian
हिन्दी,
தமிழ்,
తెలుగు,
Urdu,
ಕನ್ನಡ,
Southeast Asian
Indonesian,
Vietnamese,
Thai,
မြန်မာဘာသာ,
বাংলা
European
español,
Deutsch,
français,
Greek,
português do Brasil,
polski,
română,
русский,
Nederlands,
norsk,
svenska,
suomi,
Italian
Middle Eastern & African
عربى,
Turkish,
Persian,
Hebrew,
Afrikaans,
isiZulu,
Kiswahili,
Other
Bulgarian,
Hungarian,
Czech,
Swedish,
മലയാളം,
मराठी,
ਪੰਜਾਬੀ,
ગુજરાતી,
Portuguese,
Ukrainian