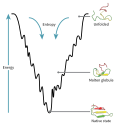Protein folding: Difference between revisions
CSV import |
CSV import |
||
| Line 49: | Line 49: | ||
{{stub}} | {{stub}} | ||
== Protein_folding == | |||
<gallery> | |||
File:Protein_folding.png|Protein folding | |||
File:Protein_structure.png|Protein structure | |||
File:Alpha_helix.png|Alpha helix | |||
File:BetaPleatedSheetProtein.png|Beta pleated sheet protein | |||
File:225_Peptide_Bond-01.jpg|Peptide bond | |||
File:Protein_folding_schematic.png|Protein folding schematic | |||
File:Molecular_Dynamics_Simulation_of_the_Hydrophobic_Solvation_of_Argon.webm|Molecular dynamics simulation of the hydrophobic solvation of argon | |||
File:PDB_1gme_EBI.jpg|PDB 1gme EBI | |||
File:X_ray_diffraction.png|X-ray diffraction | |||
File:Protein_Structural_changes_timescale_matched_with_NMR_experiments.png|Protein structural changes timescale matched with NMR experiments | |||
File:Folding_funnel_schematic.svg|Folding funnel schematic | |||
File:ACBP_MSM_from_Folding@home.tiff|ACBP MSM from Folding@home | |||
</gallery> | |||
Latest revision as of 21:08, 23 February 2025
Protein folding is the physical process by which a protein chain acquires its native 3-dimensional structure, a conformation that is usually biologically functional, in an expeditious and reproducible manner. It is the physical process by which a polypeptide folds into its characteristic and functional three-dimensional structure from a random coil.
Overview[edit]
Each protein exists as an unfolded polypeptide or random coil when translated from a sequence of mRNA to a linear chain of amino acids. This polypeptide lacks any stable (long-lasting) three-dimensional structure (the left hand side of the first figure). As the polypeptide chain is being synthesized by a ribosome, the linear chain begins to fold into its three dimensional structure. Folding begins to occur even during translation of the polypeptide chain. Amino acids interact with each other to produce a well-defined three-dimensional structure, the folded protein (the right hand side of the figure), known as the native state. The resulting three-dimensional structure is determined by the amino acid sequence or primary structure.
Protein folding and diseases[edit]
Protein misfolding is believed to be the primary cause of Alzheimer's disease, Parkinson's disease, Huntington's disease, Creutzfeldt–Jakob disease, and many other neurodegenerative disorders. Misfolded proteins can also cause cancer, cardiovascular diseases, and diabetes.
See also[edit]
- Anfinsen's dogma
- Chaperone (protein)
- Folding@home
- Levinthal's paradox
- Molecular chaperone
- Protein dynamics
- Protein structure prediction
- Protein structure prediction software
- Proteopathy
References[edit]
<references />
External links[edit]
- Protein Folding at the Protein Data Bank
|
|
|
Protein_folding[edit]
-
Protein folding
-
Protein structure
-
Alpha helix
-
Beta pleated sheet protein
-
Peptide bond
-
Protein folding schematic
-
Molecular dynamics simulation of the hydrophobic solvation of argon
-
PDB 1gme EBI
-
X-ray diffraction
-
Protein structural changes timescale matched with NMR experiments
-
Folding funnel schematic
-
ACBP MSM from Folding@home
- Protein structure
- Protein folding
- Protein biosynthesis
- Protein targeting
- Protein quaternary structure
- Protein tertiary structure
- Protein secondary structure
- Protein primary structure
- Protein conformation
- Protein dynamics
- Protein structure prediction
- Protein structure prediction software
- Proteopathy
- Protein folding diseases
- Protein misfolding diseases
- Protein folding disorders
- Protein folding problems
- Protein folding and disease
- Protein folding and neurodegenerative diseases
- Protein folding and cancer
- Protein folding and cardiovascular diseases
- Protein folding and diabetes