Oncogenomics: Difference between revisions
CSV import Tags: mobile edit mobile web edit |
CSV import |
||
| Line 39: | Line 39: | ||
{{stub}} | {{stub}} | ||
<gallery> | |||
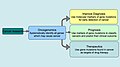
File:Overall_goals_of_oncogenomics.JPG|Overall goals of oncogenomics | |||
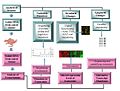
File:Current_technologies_being_used_in_Oncogenomics.jpg|Current technologies being used in Oncogenomics | |||
</gallery> | |||
Latest revision as of 01:41, 18 February 2025
Oncogenomics is a sub-field of genomics that characterizes cancer-associated genes. It focuses on genomic, epigenomic and transcript alterations in cancer.
Cancer is a genetic disease caused by accumulation of DNA mutations and epigenetic alterations leading to unrestrained cell proliferation and neoplasm formation. The goal of oncogenomics is to identify new oncogenes or tumor suppressor genes that may provide new insights into cancer diagnosis, predicting clinical outcome of cancers and new targets for cancer therapies. The success of targeted cancer therapies such as Gleevec, Herceptin and Avastin raised the hope for oncogenomics to elucidate new targets for cancer treatment.
History[edit]
The term "oncogenomics" was first coined in a paper published in 1999. The field has grown substantially since then, with major contributions from researchers in the fields of genetics, oncology, and biotechnology.
Methodology[edit]
Oncogenomics utilizes high-throughput technologies such as microarray and next-generation sequencing, and bioinformatics to understand the genetic and molecular mechanisms that drive cancer progression.
Applications[edit]
The applications of oncogenomics are numerous and include the potential for personalized cancer treatment. By understanding the specific genetic alterations that occur in individual cancers, treatments can be tailored to effectively target the unique genetic profile of each tumor.
See also[edit]
References[edit]
<references />