AlphaFold: Difference between revisions
CSV import Tags: mobile edit mobile web edit |
CSV import |
||
| Line 5: | Line 5: | ||
File:CASP_results_2020.png|CASP results 2020 | File:CASP_results_2020.png|CASP results 2020 | ||
</gallery> | </gallery> | ||
== AlphaFold == | |||
'''AlphaFold''' is an artificial intelligence (AI) program developed by [[DeepMind]], a subsidiary of [[Alphabet Inc.]], designed to predict the [[three-dimensional structure]] of [[protein]]s from their [[amino acid sequence]]. It represents a significant advancement in the field of [[computational biology]] and [[bioinformatics]]. | |||
=== Background === | |||
Proteins are complex molecules that play critical roles in biological systems. Understanding their structure is essential for insights into their function and for applications in [[drug discovery]], [[biotechnology]], and [[disease research]]. Traditionally, protein structures have been determined using experimental techniques such as [[X-ray crystallography]], [[nuclear magnetic resonance]] (NMR) spectroscopy, and [[cryo-electron microscopy]]. However, these methods can be time-consuming and expensive. | |||
=== Development === | |||
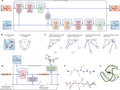
AlphaFold was developed by DeepMind as part of its efforts to apply AI to complex scientific problems. The program uses a [[deep learning]] approach, specifically a [[neural network]] architecture, to predict protein structures. It was trained on a large dataset of known protein structures and sequences, allowing it to learn patterns and relationships between sequences and their corresponding structures. | |||
=== CASP Competition === | |||
AlphaFold gained significant attention after its performance in the [[Critical Assessment of protein Structure Prediction]] (CASP) competition, a biennial event that evaluates the accuracy of protein structure prediction methods. In the CASP13 competition held in 2018, AlphaFold outperformed other methods, demonstrating its potential to revolutionize the field. In the subsequent CASP14 competition in 2020, AlphaFold achieved even greater success, with predictions that were comparable to experimental results. | |||
=== Impact === | |||
The success of AlphaFold has had a profound impact on the field of structural biology. It has accelerated research by providing accurate predictions of protein structures, which can be used to understand biological processes and develop new therapeutics. The availability of AlphaFold's predictions has also democratized access to structural information, enabling researchers worldwide to benefit from its capabilities. | |||
=== Future Directions === | |||
While AlphaFold represents a major breakthrough, there are still challenges and opportunities for further development. Future work may focus on improving the accuracy of predictions for proteins with complex structures, understanding protein dynamics, and integrating AlphaFold with other computational and experimental methods. Additionally, the principles underlying AlphaFold's success may be applied to other areas of biology and medicine. | |||
== Related pages == | |||
* [[Protein folding]] | |||
* [[Deep learning]] | |||
* [[Bioinformatics]] | |||
* [[Structural biology]] | |||
* [[DeepMind]] | |||
{{Artificial intelligence}} | |||
{{Computational biology}} | |||
[[Category:Artificial intelligence]] | |||
[[Category:Computational biology]] | |||
[[Category:Bioinformatics]] | |||
Latest revision as of 00:36, 19 February 2025
-
Protein folding figure
-
AlphaFold 2
-
Architectural details of AlphaFold 2
-
CASP results 2020
AlphaFold[edit]
AlphaFold is an artificial intelligence (AI) program developed by DeepMind, a subsidiary of Alphabet Inc., designed to predict the three-dimensional structure of proteins from their amino acid sequence. It represents a significant advancement in the field of computational biology and bioinformatics.
Background[edit]
Proteins are complex molecules that play critical roles in biological systems. Understanding their structure is essential for insights into their function and for applications in drug discovery, biotechnology, and disease research. Traditionally, protein structures have been determined using experimental techniques such as X-ray crystallography, nuclear magnetic resonance (NMR) spectroscopy, and cryo-electron microscopy. However, these methods can be time-consuming and expensive.
Development[edit]
AlphaFold was developed by DeepMind as part of its efforts to apply AI to complex scientific problems. The program uses a deep learning approach, specifically a neural network architecture, to predict protein structures. It was trained on a large dataset of known protein structures and sequences, allowing it to learn patterns and relationships between sequences and their corresponding structures.
CASP Competition[edit]
AlphaFold gained significant attention after its performance in the Critical Assessment of protein Structure Prediction (CASP) competition, a biennial event that evaluates the accuracy of protein structure prediction methods. In the CASP13 competition held in 2018, AlphaFold outperformed other methods, demonstrating its potential to revolutionize the field. In the subsequent CASP14 competition in 2020, AlphaFold achieved even greater success, with predictions that were comparable to experimental results.
Impact[edit]
The success of AlphaFold has had a profound impact on the field of structural biology. It has accelerated research by providing accurate predictions of protein structures, which can be used to understand biological processes and develop new therapeutics. The availability of AlphaFold's predictions has also democratized access to structural information, enabling researchers worldwide to benefit from its capabilities.
Future Directions[edit]
While AlphaFold represents a major breakthrough, there are still challenges and opportunities for further development. Future work may focus on improving the accuracy of predictions for proteins with complex structures, understanding protein dynamics, and integrating AlphaFold with other computational and experimental methods. Additionally, the principles underlying AlphaFold's success may be applied to other areas of biology and medicine.
Related pages[edit]
| Artificial intelligence |
|---|
|
|
| Computational science | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|