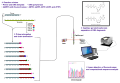Sanger sequencing: Difference between revisions
CSV import |
CSV import |
||
| Line 25: | Line 25: | ||
{{stub}} | {{stub}} | ||
== Sanger_sequencing == | |||
<gallery> | |||
File:Sanger-sequencing.svg|Sanger sequencing diagram | |||
File:Sequencing.jpg|Sequencing machine | |||
File:DNA_Sequencin_3_labeling_methods.jpg|DNA sequencing labeling methods | |||
File:Radioactive_Fluorescent_Seq.jpg|Radioactive and fluorescent sequencing | |||
File:CE_Basic.jpg|Capillary electrophoresis basics | |||
File:Sanger_sequencing_read_display.png|Sanger sequencing read display | |||
</gallery> | |||
Latest revision as of 21:12, 23 February 2025
Sanger sequencing, also known as the chain termination method, is a method of DNA sequencing first commercialized by Frederick Sanger and his colleagues in 1977. This method is based on the selective incorporation of chain-terminating dideoxynucleotides by DNA polymerase during in vitro DNA replication.
History[edit]
The Sanger sequencing method was developed in 1977 by two-time Nobel laureate Frederick Sanger and his colleagues. This method quickly replaced the then-used plus and minus method and became the most widely used DNA sequencing method for the next 40 years.
Method[edit]
The Sanger sequencing method involves four separate DNA synthesis reactions. Each reaction includes a DNA template, a DNA primer, the four deoxyribonucleotide (dNTP) bases, DNA polymerase, and a small amount of one of the four dideoxynucleotide (ddNTP) bases. The ddNTPs cause DNA synthesis to stop, as they lack the 3'-OH group required for the formation of a phosphodiester bond between two nucleotides, causing DNA polymerase to cease extension of DNA.
Applications[edit]
Sanger sequencing is used in a wide range of applications, including gene sequencing, mutation detection, and genetic disease diagnosis. It is also used in research for studying genetic disorders, cancer, and infectious diseases.
Limitations[edit]
While Sanger sequencing is a reliable and accurate method for DNA sequencing, it has several limitations. These include the inability to sequence very long DNA molecules, the requirement for a large amount of purified DNA, and the relatively high cost and time-consuming nature of the method.